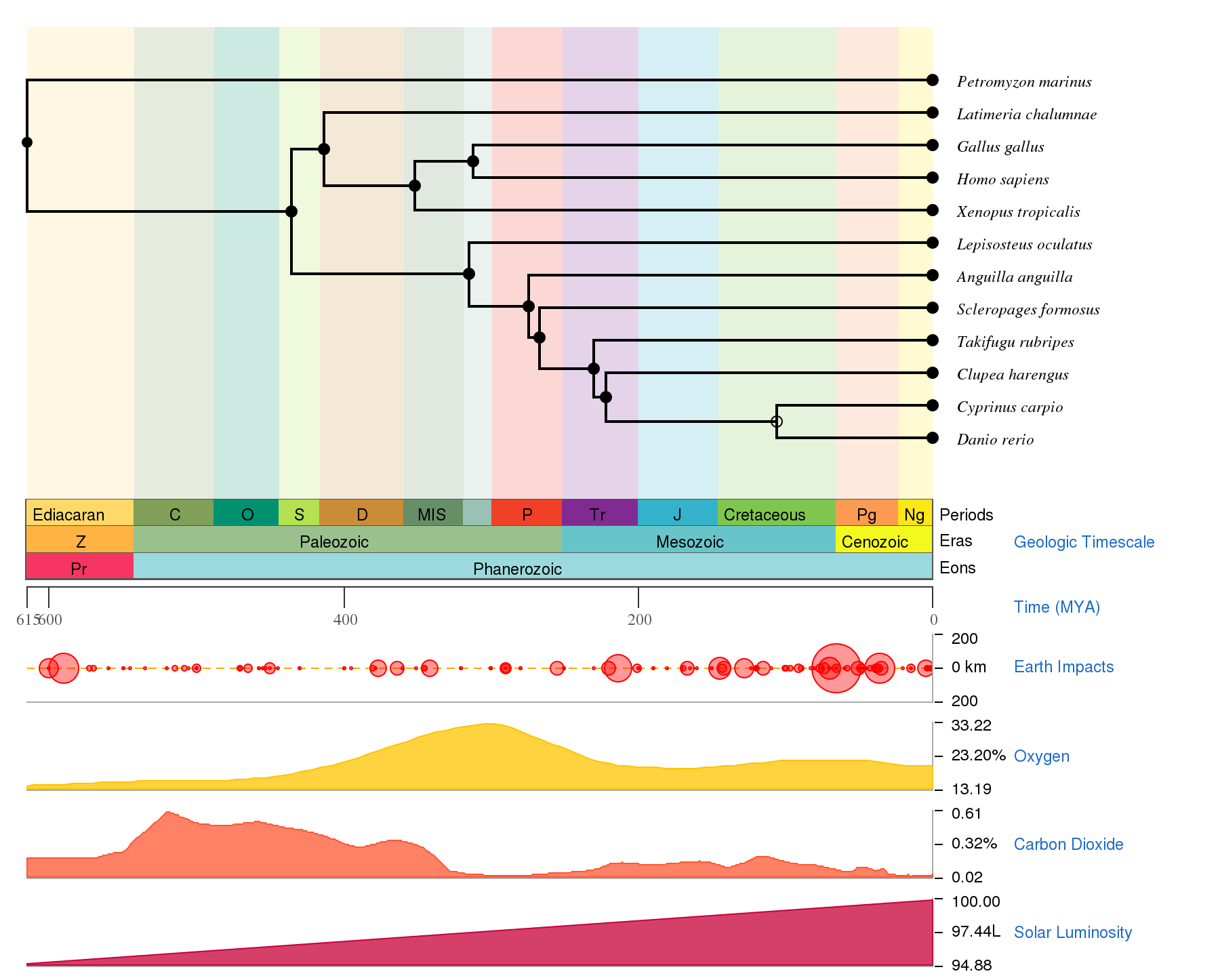
A tree 400MYA in the making: choosing species for a phylogenetic analysis between zebrafish and human genes
As of June of 2018, this is the list of species that have a sequenced genome (at least partially) and I use in phylogenetic and reconciliation analysis focused on investigating genetic evolution between zerbafish and mammals:
One of the analysis that I like to do when trying to understand the function of a specific gene is to recapitulate its evolution since the last common ancestor between zebrafish and humans. This brings a problem: from which species do I have to search and download data to convincingly fill the gap on this divergence times? How many data points do we need to bridge a 400 million year gap, including (at least) one full whole-genome duplication?
Species tree created with TimeTree
While there are countless publications with phylogenetic trees made with the sequences from zebrafish, mouse, humans, and either chicken or frogs, I found these datasets to be skewed towards tetrapods, with insufficient fish species coverage and usually lead to wrong conclusions.
As of 2018, this is the list of species that have a sequenced genome (at least partially) and I use in phylogenetic and reconciliation analysis focused on investigating genetic evolution between zebrafish and mammals:
| Last common taxonomy | Species |
|---|---|
| Vertebrata | Lampreys |
| Gnathostomata | Sharks |
| Telestomi | |
| Eutelestomi | Latimeria, lungfishes & Tetrapods |
| Actinopterygii | |
| Actinopteri | |
| Neopterygii | Lepisosteus oculatus |
| Teleostei | Anguilla anguilla |
| Osteoglossocephalai | Scleropages formosus, Paramormyrops kingsleyae |
| Clupeocephala | Great majority of fishes with sequenced genome: Takifugu, Tetraodon, Oryzias, Gasterosteus, Peocilia, Xiphophorus, ... |
| Otomorpha | Clupea harengus |
| Ostariophysi | |
| Otophysi | Astyanax mexicanus, Electrophorus electricus, Ictalurus punctatus, Pygocentrus nattereri |
| Cypriniphysae | |
| Cypriniformes | |
| Cyprinoidea | |
| Cyprinidae | Cyprinus carpio, Squalius pyrenaicus, Sinocyclocheilus rhinocerous, anshuiensis, and grahami, Leuciscus waleckii, Pimephales promelas |
| Danio | Danio rerio |
I often use sequences from lampreys and sharks to root the trees; the coealacanth and lungfishes help in joining the tetrapod and often diverging fish lineages; and the spotted gar is the only non-teleost fish with sequenced genome that I know of, which is useful to pinpoint duplications emerging from the whole genome duplication.