Preparing a zebrafish RNAseq for GSEA
Recently I had to annotate a rnaseq analysis that was only mapped to Ensembl IDs for an enrichment analysis in GSEA and this is the way in which I am getting the most insightful results on my research.
Recently I had to annotate a rnaseq analysis that was only mapped to Ensembl IDs for an enrichment analysis in GSEA. That meant getting the maximum number of zebrafish Ensembl gene IDs to its human homolog/ortholog (you know the drill with zebrafish) Gene Symbols. So I spent some time analysing different mapping ways from to lose the least amount of information possible while still preserving the variability of zebrafish duplicates.
Some options that I tried:
- Use the GSEA-provided zebrafish.chip
- Simply get all genes to caps and use only the direct matches between zebrafish and human genes
- Get homologs through:
3.1. Biodbnet
3.2. The RAnnotationDBipackage (org.Dr.eg.dbin this case)
3.3.biomaRt - A bunch of other crazy stuff
In the end, I found that using a two-step process would yield the best results for the time spent:
- Map the Ensembl IDs to ZFIN IDs
- Use the ZFIN database as a chip for the conversion
1. From zebrafish Ensembl Gene IDs to ZFIN IDs
Thankfully, the mapping from Ensembl IDs to ZFIN IDs is pretty easy and straightforward. Here are two R functions to get the job done through the biomaRt and org.Dr.eg.db packages:
# Function txdbAnnot.
# Input: a list of Ensembl Gene IDs.
# Return: an annotated dataframe.
txdbAnnot <- function(listOfIDS,
attributes = attributes,
keys = keys) {
library("org.Dr.eg.db")
# print("Annotating using org.Dr.eg.db")
txdbAnnotDF <- select(org.Dr.eg.db,
keys=listOfIDS,
columns=attributes,
keytype=keys,
multiVals="first")
return(txdbAnnotDF)
}
bioMartConversion <- function(listOfIDS,
attributes = attributes,
filters = filters) {
library("biomaRt")
# print("Annotating using BioMaRt")
zfishMart <- useMart("ensembl", dataset="drerio_gene_ensembl")
geneAnnot <- getBM(attributes = attributes,
filters = filters,
values = listOfIDS,
mart = zfishMart)
return(geneAnnot)
}
# Function deleDuplicatesDataFrame.
# Input: a dataframe and a column with duplicates.
# Output: the dataframe without the duplicates.
deleteDuplicatesDataFrame <- function(df, col) {
dup.idx <- which(duplicated(df[col]))
return(df[-dup.idx,])
}
And then annotate using one of the methods:
# You can use the call to annotate with other IDs as
# attributes <- c("ENTREZID", "SYMBOL", "ZFIN", "GENENAME")
zfinTxdb <- txdbAnnot(listOfIDS,
attributes = "ZFIN",
keys = "ENSEMBL")
# Clean up the duplicates
zfinTxdb <- deleteDuplicatesDataFrame(zfinTxdb, "ZFIN")
or
# You can use the call to annotate with other IDs as
# attributes <- c("ensembl_gene_id", "entrezgene", "external_gene_name",
"zfin_id_id", "description")
zfinBiomaRt <- bioMartConversion(listOfIDS,
attributes = c("ensembl_gene_id","zfin_id_id"),
filters = "ensembl_gene_id")
# Clean up the duplicates
zfinBiomaRt <- deleteDuplicatesDataFrame(zfinBiomaRt, "ensembl_gene_id")
I personally prefer using AnnotationDBI because it's faster.
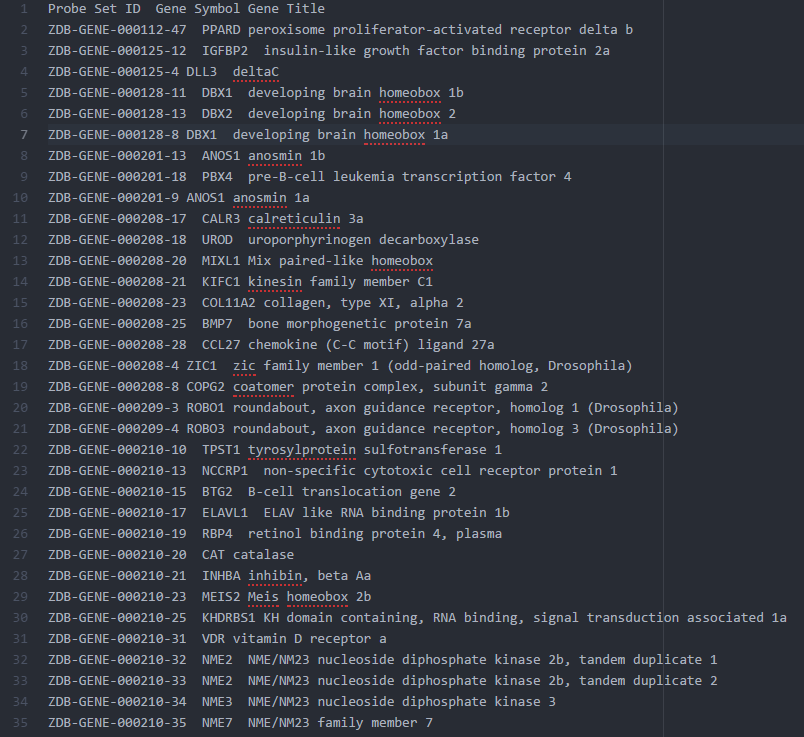
2. The ZFIN.chip for GSEA
ZFIN provides data dumps for most of its data, and one really useful table is the Human and Zebrafish Orthology table. You can find it at the downloads page.
Just create a mapping chip without duplicates using the columns ZFIN ID as Probe, Human gene Symbol as Gene Symbol, and the zebrafish gene name as Gene Title. That way the 1-to-many orthologs can be treated as different (technically, as different probes for the same gene) but we don't lose info or pick just one randomly.
That's it. I know that it's not perfect, but this is the way in which I am getting the most insightful results on my research.
What are your thoughts? Get in touch and discuss it!